1 Introduction
Categorical data are prevalent in all areas, including economics, marketing, finance, psychology, and clinical studies. To analyze categorical data, the probit or logit models are often used to make inferences. To perform model assessment and comparison, researchers often rely on goodness-of-fit measures, such as ${R^{2}}$ (also known as the coefficient of determination). For example, the ordinary least square (OLS) ${R^{2}}$ is one of the most extensively used goodness-of-fit measures for linear models in continuous data analysis. For categorical data analysis, however, there is no such universally adopted ${R^{2}}$ measure [19, 39]. Continuous efforts have been made to develop sensible ${R^{2}}$ measures for probit/logistic models, and more generally, generalized linear models [29, 30, 14, 11, 22, 42, 27, 21, 26]. Among the existing ${R^{2}}$ measures, McKelvey-Zavoina’s ${R_{MZ}^{2}}$ [30] and McFadden’s ${R^{2}}$ [29] are probably the most well-known and widely used in domain research [19, 39]. But as demonstrated in [26], Mckelvey-Zavoina’s ${R_{MZ}^{2}}$ does not hold monotonicity, which means a larger model may have a smaller ${R_{MZ}^{2}}$. This serious defection of ${R_{MZ}^{2}}$ may be misleading in practice and misguide the model-building process. On the other hand, McFadden’s ${R^{2}}$ relies on the ratio of likelihoods, and it does not preserve the interpretation of explained variance. Neither of these two ${R^{2}}$ measures meets all of the three criteria considered in [26]:
[26] proposed a so-called surrogate ${R^{2}}$ that satisfies all three criteria for probit models. This surrogate ${R^{2}}$ uses the notion of surrogacy, namely, generating a continuous response S and using it as a surrogate for the original categorical response Y [24, 25, 8, 18, 23]. In the context of probit analysis, [26] used the truncated distributions induced by the latent variable structure to generate a surrogate response S. This surrogate response S is then regressed on explanatory variables through a linear model. The OLS ${R^{2}}$ of this linear model is used as a surrogate ${R^{2}}$ for the original probit model. This surrogate ${R^{2}}$ meets all three criteria (C1)–(C3).
The goals of this paper are (i) developing an R package to implement [26]’s method; (ii) demonstrating how this new package can be used jointly with other existing R packages for variable selection and model diagnostics in the model building process; and (iii) illustrating how this package can be used to compare different empirical models trained from two different samples (a.k.a. comparability) in real data analysis.
Specifically, we first develop an R package to implement the surrogate ${R^{2}}$ method for probit/logistic regression models. This package contains the R functions for generating the point and interval estimates of the surrogate ${R^{2}}$ measure. The point and interval estimates allow researchers and practitioners to evaluate the model’s overall goodness of fit and understand its uncertainty. In addition, we develop an R function that calculates the percentage contribution of each variable to the overall surrogate ${R^{2}}$. This percentage reflects each variable’s contribution to the model’s overall explanatory power. Based on the contribution’s relative size, our R function provides an “importance” ranking of all the explanatory variables.
Second, to provide practical guidance for categorical data modeling, we use the developed R package to demonstrate how it can be used jointly with other R packages developed for variable screening/selection and model diagnostics ($\mathbf{leaps}$ [28], step() function from the R core, glmnet [17], ordinalNet [40], ncvreg [5], grpreg [6], SIS [34], sure [18], $\mathbf{PAsso}$ [44]). In particular, we recommend a workflow that consists of three steps: variable screening/selection, model diagnostics, and goodness-of-fit analysis. The workflow is illustrated in the analysis of wine-tasting preference datasets.
Third, the comparability of the surrogate ${R^{2}}$ across different samples and/or models allows us to compare goodness-of-fit analysis from similar studies. The comparison can lead to additional scientific and business insights which may be useful for decision making. To illustrate this, we conduct goodness-of-fit analysis separately for the red wine and white wine samples to demonstrate the comparability of the surrogate ${R^{2}}$. Our analysis result reveals that (i) the same set of explanatory variables has different explanatory power for red wine and white wine (43.8% versus 31.0%), and (ii) the importance ranking of the explanatory variable (in terms of their contribution to the surrogate ${R^{2}}$) is different between red wine and white wine.
Our $\mathbf{SurrogateRsq}$ package has broad applicability. It is compatible with the following R functions that can fit probit/logistic regression models for a binary or ordinal response: glm() in the R core, polr() in the $\mathbf{MASS}$ package [33], clm() in the $\mathbf{ordinal}$ package [9], and vglm() in the $\mathbf{VGAM}$ package [41], although we only demonstrate the functions of $\mathbf{SurrogateRsq}$ package through ordinal probit regression models using plor() in this paper. More examples and details can be found on the website: https://xiaorui.site/SurrogateRsq/.
2 Review of the Surrogate ${R^{2}}$
We briefly review the surrogate ${R^{2}}$ measure in the study of [26]. For the model setting, we consider a probit/logit model with a set of explanatory variables. The categorical response is either a binary or ordinal variable Y that has J categories $\{1,2,\dots ,J\}$, with the order $1\lt 2\lt \cdots \lt J$,
where $-\infty \lt {\alpha _{1}}\lt \cdots \lt {\alpha _{J}}\lt +\infty $. The link function $G(\cdot )$ can be a probit ($G(\cdot )=\Phi (\cdot )$) link or a logit ($G(\eta )=1/(1+{e^{-\eta }})$). Each generic symbol of {${X_{1}},\dots {X_{p}}$} in Model (2.1) can represent a single variable of interest, a high-order term (e.g., ${X^{2}}$), or an interaction term between X and another variable. It is well-known that an equivalent way to express Model (2.1) is through a latent variable. For example, if the link is probit, the latent variable has the following form with a normally distributed ϵ:
(2.1)
\[ \Pr \{Y\le j\}=G\{{\alpha _{j}}-({\beta _{1}}{X_{1}}+\cdots +{\beta _{l}}{X_{p}})\},\hspace{1em}j=1,\dots ,J,\]The categorical response Y can be viewed as generated from censoring the continuous latent variable Z in the following way:
\[ Y=\left\{\begin{array}{l@{\hskip10.0pt}l}1& \hspace{1em}\text{if}\hspace{2.5pt}-\infty \lt Z\le {\alpha _{1}}+{\alpha _{1}},\\ {} 2& \hspace{1em}\text{if}\hspace{2.5pt}{\alpha _{1}}+{\alpha _{1}}\lt Z\le {\alpha _{2}}+{\alpha _{1}},\\ {} \cdots \\ {} J& \hspace{1em}\text{if}\hspace{2.5pt}{\alpha _{J-1}}+{\alpha _{1}}\lt Z\lt +\infty .\end{array}\right.\]
To construct a goodness-of-fit ${R^{2}}$, [26] adopted the surrogate approach proposed by [24]. The idea of the surrogate approach is to simulate a continuous variable and use it as a surrogate for the original categorical variable in the analysis [24, 25, 8]. In the context of probit models, [26] proposed to generate a surrogate response variable using the following truncated conditional distribution:
\[ S\sim \left\{\begin{array}{l@{\hskip10.0pt}l}Z\mid -\infty \lt Z\le {\alpha _{1}}+{\alpha _{1}}& \hspace{1em}\text{if}\hspace{2.5pt}Y=1,\\ {} Z\mid {\alpha _{1}}+{\alpha _{1}}\lt Z\le {\alpha _{2}}+{\alpha _{1}}& \hspace{1em}\text{if}\hspace{2.5pt}Y=2,\\ {} \cdots \\ {} Z\mid {\alpha _{J-1}}+{\alpha _{1}}\lt Z\lt +\infty & \hspace{1em}\text{if}\hspace{2.5pt}Y=J.\end{array}\right.\]
[26] proposed to regress the surrogate response S on {${X_{1}},\dots ,{X_{p}}$} using a linear model below:
Their approach used the OLS ${R^{2}}$ measure of this linear model as a surrogate ${R^{2}}$ for Model (2.1):
(2.2)
\[ S={\alpha _{1}}+{\beta _{1}}{X_{1}}+\cdots +{\beta _{p}}{X_{p}}+\epsilon ,\hspace{1em}\epsilon \sim N(0,1).\][26] showed that the surrogate ${R_{(S)}^{2}}$ measure has three desirable properties. First, it approximates the OLS ${R^{2}}$ calculated using the latent continuous outcome Z. This property enables us to compare surrogate ${R^{2}}$’s and OLS ${R^{2}}$’s across different models and samples that address the same scientific question. Second, as it is the OLS ${R^{2}}$ calculated using the continuous surrogate response S, the surrogate ${R_{(S)}^{2}}$ has the interpretation of the explained proportion of variance. It measures the explained proportion of the variance of the surrogate response S through the linear model. This explained proportion of variance implies the explanatory power of all the features in the fitted model. Third, the surrogate ${R_{(S)}^{2}}$ maintains monotonicity between nested models, which makes it suitable for comparing the relative explanatory power of different models. In contrast, the well-known McFadden’s ${R^{2}}$ does not preserve the first two properties of the surrogate ${R_{(S)}^{2}}$. McFadden’s ${R^{2}}$ relies on the ratio of likelihoods, so it neither approximates the OLS ${R^{2}}$ nor preserves the interpretation of explained variance. On the other hand, [26] showed that McKelvey-Zavoina’s ${R_{MZ}^{2}}$ did not necessarily maintain monotonicity between nested models. This serious issue may make McKelvey-Zavoina’s ${R_{MZ}^{2}}$ an unsuitable tool for measuring the goodness of fit.
To make inferences for the surrogate ${R_{(S)}^{2}}$, [26] provided procedures to produce point and interval estimates. Since the surrogate response S is obtained through simulation, [26] used a multiple-sampling scheme to “stabilize” the point estimate. They also provided an implementation to produce an interval estimate with a $95\% $ confidence level. This confidence interval is constructed through a bootstrap-based pseudo algorithm. When the sample size is large (e.g., $n=2000$), [26]’s numerical studies show that the interval measure of the surrogate ${R_{(S)}^{2}}$ can approximate the nominal coverage probability.
It is also worth noting that [26]’s method requires a full model. This paper will illustrate how to use existing tools, such as variable selection and model diagnostics, to initiate a full model. The full model is used to generate a common surrogate response S, which is then used to calculate the surrogate ${R_{(S)}^{2}}$’s of whatever reduced models. We will demonstrate how to carry it out in a real data analysis presented in Section 5.
3 Main Functions of the $\textbf{SurrogateRsq}$ Package
We develop an R package $\mathbf{SurrogateRsq}$ for goodness-of-fit analysis of probit models [43]. This package contains functions to provide (i) a point estimate of the surrogate ${R^{2}}$; (ii) an interval estimate of the surrogate ${R^{2}}$; (iii) an importance ranking of explanatory variables based on their contributions to the total surrogate ${R^{2}}$ of the full model; and (iv) other existing ${R^{2}}$ measures in the literature. In this section, we explicitly explain the inputs and outputs of these functions. In the next two sections, we will demonstrate the use of these functions through a recommended workflow and real data examples.
-
1. surr_rsq: a function for producing a point estimate of the surrogate ${R_{(S)}^{2}}$ for a user-specified model. It requires three inputs: a reduced model, a full model, and a dataset. This function generates an S3 object of the class “surr_rsq”. Other functions in this package can directly call this S3 object. The details of the three inputs are as follows:
-
• model: a model to be evaluated for the goodness of fit. Our implementation supports a few popular classes of objects. They are the probit model from the glm function in the R core stats package, the ordered probit model generated from the plor function in the MASS package, clm() in the ordinal package, and vglm() in the VGAM package.
-
• avg.num: an optional input that specifies the numbers of simulations used in multiple sampling. The default value is 30. The surrogate ${R_{(S)}^{2}}$ is calculated using the simulated surrogate response S. A multiple-sampling scheme can be used to “stabilize” the point estimate of ${R_{(S)}^{2}}$ by using the average of multiple ${R_{(S)}^{2}}$’s values.
-
• asym: an optional logical argument that specifies whether to use the asymptotic version of the surrogate R-squared. The default value is FALSE. If TRUE, we calculate the surrogate ${R^{2}}$ using the asymptotic formula on page 208 of the paper by [26]. More details are provided in that paper. This approach avoids calculating the average of multiple ${R_{(S)}^{2}}$ in the above argument.
-
-
2. surr_rsq_ci: a function for generating an interval measure of the surrogate ${R^{2}}$ with the designated confidence level. This interval accounts for and reflects the uncertainty in the ${R^{2}}$ statistic. This function requires three inputs:
-
• object: an object generated from the previous surr_rsq function.
-
• alpha: the value of alpha determines the confidence level of the interval, namely, $100(1-\alpha )\% $. The default value of alpha is 0.05.
-
• B: the number of bootstrap replications. The default value of B is 2000. The confidence interval is derived from a bootstrap distribution for ${R_{(S)}^{2}}$. See the section of “Inference by Multiple Sampling” in [26].
-
• asym: an optional logical argument that specifies whether to use the asymptotic version of the surrogate R-squared. The default value is FALSE.
-
• parallel: an optional logical argument that controls parallel computing using the foreach [1]. The default value is FALSE. If TRUE, the parallel clusters need to be registered through registerDoParallel() or registerDoSNOW() beforehand.
-
Figure 1
An illustration of a workflow for modeling categorical data. Grey boxes show statistical analysis steps that should be carried out before our goodness-of-fit analysis. Light blue boxes contain the main functions in the SurrogateRsq package. Orange boxes highlight inference outcomes produced by our SurrogateRsq package.
-
3. surr_rsq_rank: a function to give ranks of explanatory variables based on their contributions to the overall surrogate ${R^{2}}$. The rank is based on the variance contribution of each variable. Specifically, it calculates the reduction of the surrogate ${R_{(S)}^{2}}$ of the model that removes each variable one at a time. The rank is then determined according to the reduction, which indicates the importance of each variable relevant to others. In addition to the ranks, the output table includes the ${R^{2}}$ reduction and its percentage in reference to the total surrogate ${R^{2}}$ of the full model. The function only requires the object input. It is a generated object from the surr_rsq function. The optional avg.num argument is the same as the one in the surr_rsq function, and the option var.set is explained below.
-
• object: an object generated from the previous surr_rsq function.
-
• var.set: an optional argument that allows users to examine the contribution of a set of variables, as a whole, to the total surrogate ${R^{2}}$. If not specified, the function calculates the goodness-of-fit contributions to the overall surrogate ${R^{2}}$ for individual variables.
-
4 Using R Packages for Categorical Data Modeling: A Workflow
In empirical studies, goodness-of-fit analysis should be used jointly with other statistical tools, such as variable screening/selection and model diagnostics, in the model-building and refining process. In this section, we discuss how to follow the workflow in Figure 1 to carry out statistical modeling for categorical data. We also discuss how to use the $\mathbf{SurrogateRsq}$ package with other existing R packages to implement this workflow. As [26]’s method requires a full model, researchers and practitioners can also follow the process in Figure 1 to initiate a full model to facilitate goodness-of-fit analysis.
-
1. In Step-0, we can use the AIC/BIC/LASSO or any other variable selection methods deemed appropriate to trim or prune the set of explanatory variables to a “manageable” size (e.g., less than 20). The goal is to eliminate irrelevant variables so researchers can better investigate the model structure and assessment. The variable selection techniques have been studied extensively in the literature. Specifically, one can implement (i) the best subset selection using the function regsubsets() in the leaps package; (ii) the forward/backward/stepwise selection using the function step() in the R core; (iii) the shrinkage methods including the (adaptive) LASSO in the glmnet package; (iv) the regularized ordinal regression model with an elastic net penalty in the ordinalNet package; and (v) the penalized regression models with minimax concave penalty (MCP) or smoothly clipped absolute deviation (SCAD) penalty in the ncvreg package [36, 46, 45, 35, 40]. When the dimension is ultrahigh, the sure independence screening method can be applied through the SIS package [16]. When the variables are grouped, one can apply the group selection methods including the group lasso, group MCP, and group SCAD through the grpreg package [6]. In some cases, Step-0 may be skipped if the experiment only involves a (small) set of controlled variables. In these cases, the controlled variables should be modeled regardless of statistical significance or predictive power. We limit our discussion here because our focus is on goodness-of-fit analysis.
-
2. In Step-1, we can use diagnostic tools to inspect the model passed from Step-0, adjust its functional form, and add additional elements if needed (e.g., higher-order or interaction terms). For categorical data, we can use the function autoplot.resid() in the sure package [24, 18] to generate three types of diagnostic plots: residual Q-Q plot, residual-vs-covariate plot, and residual-vs-fitted plots. These plots can be used to visualize the discrepancy between the working model and the “true” model. Similar plots can be produced using the function diagnostic.plot() in the PAsso package [44]. These diagnostic plots provide practitioners insights on how to refine the model by possibly transforming the regression form or adding higher-order terms. At the end of this diagnosing and refining process, we expect to have a full model (${\mathcal{M}_{full}}$) for subsequent inferences including goodness-of-fit analysis.
-
3. In Step-2, we can use the functions developed in our SurrogateRsq package to examine the goodness of fit of the full model ${\mathcal{M}_{full}}$ and various reduced models of interest. Specifically, we can produce the point and interval estimates of the surrogate ${R^{2}}$ by using the functions surr_rsq() and surr_rsq_ci(). In addition, we can quantify the contribution of each individual variable to the overall surrogate ${R^{2}}$ by using the function surr_rsq_rank(). Based on the percentage contribution, the function surr_rsq_rank() also provides ranks of the explanatory variables to show their relative importance. In the following section, we will show in a case study how our package can help us understand the relative importance of explanatory variables and compare the results across different samples. The “comparability” across different samples and/or models is an appealing feature of the surrogate ${R^{2}}$, which will be discussed in detail along with the R implementation.
5 Analysis of the Wine Rating Data: A Demonstration
In this section, we demonstrate how to use our SurrogateRsq package, coupled with R packages for model selection and diagnostics, to carry out statistical analysis of the wine rating data. A critical problem in wine analysis is to understand how the physicochemical properties of wines may influence human tasting preferences [10]. For this purpose, [10] collected a dataset that contains wine ratings for 1599 red wine samples and 4898 white wine samples. The response variable, wine ratings, is measured on an ordinal scale ranging from 0 (very bad) to 10 (excellent). The explanatory variables are 11 physicochemical features, including alcohol, sulphates, acidity, dioxide, pH, and others.
Our analysis of the wine rating data follows the workflow discussed in Section 4. Specifically, in Section 5.1, we initiate a full model using several R packages for variable selection and model diagnostics. In Section 5.2, we use our SurrogateRsq package to evaluate (i) the goodness-of-fit of the full model and several reduced models; (ii) the contribution of each individual variable to the overall ${R^{2}}$; and (iii) the difference between the red wine and white wine in terms of how physicochemical features may influence human tasting differently.
5.1 Initiating a Full Model Using Variable Selection and Model Diagnostics
To start, we use the function polr() to fit a probit model to the red wine sample using all 11 explanatory variables. This “naive” model has identified three explanatory variables that are insignificant: they are fixed.acidity, citric.acid, and residual.sugar.
5.1.1 Variable Selection
As the number of explanatory variables is small, we use the exhaustive search method to select variables.
Figure 2 plots the exhaustive search selection results based on the BIC. Each row in the plot represents a model that has been trained with the variables highlighted in black color. The top row is the selected model with the smallest BIC value. This model does not select fixed.acidity, citric.acid, residual.sugar, and density. Note that the first three are not significant. We will perform diagnostics on this model in the subsection that follows.
We remark that if the number of explanatory variables is (moderately) large, we can use the step-wise selection method or regularization methods (e.g., with an L1, elastic net, minimax concave, or SCAD penalty). An example code is attached in the supplementary materials.
5.1.2 Model Diagnostics
We conduct diagnostics of the model with variables selected in the preview step. For this purpose, we use surrogate residuals [24], which can be implemented by the function autoplot.resid() in the package sure [18] or the function diagnostic.plot() in the package PAsso [44]. The code below produces residual-vs-covariate plots for the object select_model by specifying the output = "covariate".
Figure 3
Plots of surrogate residual versus sulphates for (a) the model with a linear term of sulphates; (b) the model with an additional quadratic term of sulphates; and (c) the model with an additional cubic term of $sulphates$. The solid red curves are LOESS curves.
Among all the residual-vs-covariate plots, we find that the residual-vs-sulphates plot in Figure 3(a) shows an inverted U-shape pattern, which suggests a missing quadratic term of sulphates. We update the model by adding a squared term $I({\mathtt{sulphates}^{2}})$ to the object select_model and run model diagnostics again using the code below. Figure 3(b) shows that the plot for sulphates still exhibits a nonlinear pattern. We therefore add a cubic term $I({\mathtt{sulphates}^{3}})$ to the model. The LOESS curve in the updated plot in Figure 3(c) turns out to be flat. We use this model as our full model (${\mathcal{M}_{full}}$).
Table 1
Model development for the red wine by variable selection and model diagnostics.
| Dependent variable: quality | ||||
| Model | Naive | Selected | + sulphates${^{2}}$ | + sulphates${^{3}}$ |
| full model ${\mathcal{M}_{full}}$ | ||||
| fixed.acidity | 0.026 | |||
| (0.028) | ||||
| volatile.acidity | −1.868${^{\ast \ast \ast }}$ | −1.722${^{\ast \ast \ast }}$ | −1.534${^{\ast \ast \ast }}$ | −1.491${^{\ast \ast \ast }}$ |
| (0.213) | (0.180) | (0.183) | (0.183) | |
| citric.acid | −0.337 | |||
| (0.256) | ||||
| residual.sugar | 0.011 | |||
| (0.021) | ||||
| chlorides | −3.234${^{\ast \ast \ast }}$ | −3.488${^{\ast \ast \ast }}$ | −2.965${^{\ast \ast \ast }}$ | −2.604${^{\ast \ast \ast }}$ |
| (0.733) | (0.699) | (0.707) | (0.715) | |
| free.sulfur.dioxide | 0.010${^{\ast \ast \ast }}$ | 0.011${^{\ast \ast \ast }}$ | 0.010${^{\ast \ast }}$ | 0.010${^{\ast \ast }}$ |
| (0.004) | (0.004) | (0.004) | (0.004) | |
| total.sulfur.dioxide | −0.007${^{\ast \ast \ast }}$ | −0.008${^{\ast \ast \ast }}$ | −0.007${^{\ast \ast \ast }}$ | −0.007${^{\ast \ast \ast }}$ |
| (0.001) | (0.001) | (0.001) | (0.001) | |
| density | −6.679${^{\ast \ast \ast }}$ | |||
| (0.538) | ||||
| pH | −0.754${^{\ast \ast \ast }}$ | −0.780${^{\ast \ast \ast }}$ | −0.969${^{\ast \ast \ast }}$ | −1.028${^{\ast \ast \ast }}$ |
| (0.277) | (0.205) | (0.208) | (0.209) | |
| sulphates | 1.589${^{\ast \ast \ast }}$ | 1.570${^{\ast \ast \ast }}$ | 5.937${^{\ast \ast \ast }}$ | 15.147${^{\ast \ast \ast }}$ |
| (0.195) | (0.193) | (0.678) | (2.591) | |
| sulphates${^{2}}$ | −2.515${^{\ast \ast \ast }}$ | −12.397${^{\ast \ast \ast }}$ | ||
| (0.374) | (2.707) | |||
| sulphates${^{3}}$ | 3.092${^{\ast \ast \ast }}$ | |||
| (0.839) | ||||
| alcohol | 0.481${^{\ast \ast \ast }}$ | 0.479${^{\ast \ast \ast }}$ | 0.475${^{\ast \ast \ast }}$ | 0.472${^{\ast \ast \ast }}$ |
| (0.032) | (0.031) | (0.031) | (0.031) | |
Note: ${^{\ast }}$p<0.1${;^{\ast \ast }}$p<0.05${;^{\ast \ast \ast }}$p<0.01
Table 1 summarizes the model fitting results for the naive model and models progressively trained in the procedures of variable selection and model diagnostics. Compared to the naive model, the “Selected” column basically removes density and three non-significant variables, which results in a lower BIC value. The last two columns of Table 1 confirm the statistical significance of both the squared and cubic terms of sulphates, which are identified and added in the model diagnostics procedure. The model presented in the last column will be used as the full model ${\mathcal{M}_{full}}$ in our goodness-of-fit assessment in the next subsection.
5.2 Goodness-of-Fit Analysis and Its Extended Utility
In this subsection, we use our developed $\mathbf{SurrogateRsq}$ package to illustrate how to use the surrogate ${R^{2}}$ to (i) assess the goodness-of-fit of the full model and reduced models; (ii) rank exploratory variables based on their contributions to ${R^{2}}$; and (iii) compare the goodness of fit across multiple samples and/or models.
5.2.1 Surrogate ${R^{2}}$ for the Full Model
First of all, we use the function surr_rsq to calculate the surrogate ${R^{2}}$ of the full model ${\mathcal{M}_{full}}$ identified in the previous subsection. To do so, in the code below we set the arguments model and full_model to be the same as ${\mathcal{M}_{full}}$. We use 30 as the number of simulations for multiple sampling. The purpose of performing multiple sampling is to “stabilize” the point estimate of ${R^{2}}$ [26].
This function provides a point estimate of the surrogate ${R^{2}}$ of the full model. The value 0.439 implies 43.9% of the variance of the surrogate response S can be explained by the seven explanatory variables and two nonlinear terms of sulphates.
5.2.2 Surrogate ${R^{2}}$ for a Reduced Model
We can also use the same function surr_rsq to calculate the surrogate ${R^{2}}$ of a reduce model. For example, to evaluate the goodness of fit of the model without high-order terms of sulphates, we simply need to change the model argument to be the reduced model select_model as shown in the code below. The specification of the full model is still required in the code, and such a full model should be common to all the reduced models to be compared. This is a way to eliminate the non-monotonicity issue as seen in Mckelvey-Zavoina’s ${R_{MZ}^{2}}$ [26].
The result shows that the surrogate ${R^{2}}$ has been reduced to 0.41 if the squared and cubic terms of sulphates are removed from the model. This means that the high-order terms of sulphates constitute 6.60% of the total surrogate ${R^{2}}$.
5.2.3 Confidence Interval for the Surrogate ${R^{2}}$
The package $\mathbf{SurrogateRsq}$ allows us to produce a confidence interval for the surrogate ${R^{2}}$ using the function surr_rsq_ci. This function can directly use the object surr_obj_mod_full created earlier as the input of the object argument. In the code below, we set the significance level alpha = 0.05 to produce a 95% confidence interval and the number of bootstrap repetitions to be 2000. The output is a table with the lower and upper bounds of the confidence interval. For the full model ${\mathcal{M}_{full}}$, the 95% confidence interval of the surrogate ${R^{2}}$ is $[0.402,0.485]$. The tightness of this interval implies that the uncertainty of the ${R^{2}}$ inference is low. 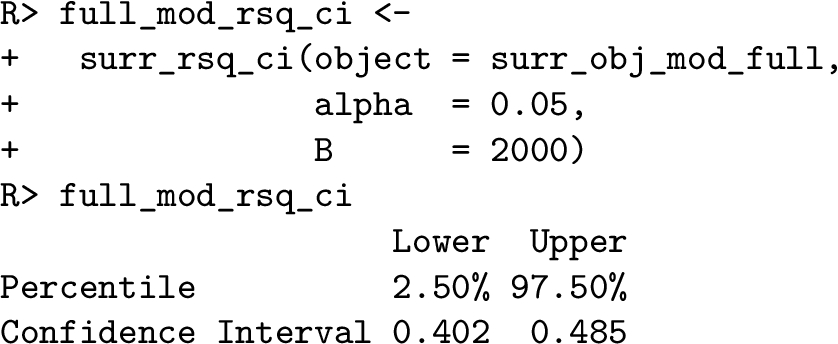
5.2.4 Importance Ranking of Explanatory Variables
We apply the function surr_rsq_rank() to examine the contribution of each individual variable to the overall surrogate ${R^{2}}$, which in turn produces a table of importance ranking. In the code below, we set the object argument as the object surr_obj_mod_full created earlier to examine the relative contribution of the variables in the full model. The output table shows (i) the surrogate ${R^{2}}$ for the model that removes an explanatory variable one at a time; (ii) the reduction of the ${R^{2}}$ after removing such a variable; (iii) the percentage contribution of this variable to the total surrogate ${R^{2}}$; and (iv) the rank of the variable by its percentage contribution. In the table below, we observe that the variable alcohol is ranked at the top as it explains 25.80% of the total surrogate ${R^{2}}$. It is followed by volatile.acidity (7.12%), total.sulfur.dioxide (3.52%), and sulphates (3.13%). The rest of the explanatory variables contribute less than 3% to the total surrogate ${R^{2}}$.
In the ranking table above, the contributions of sulphates and its higher order terms ${\mathtt{sulphates}^{2}}$ and ${\mathtt{sulphates}^{3}}$ to the surrogate ${R^{2}}$ are evaluated separately. This is the default setting of the function surr_rsq_rank() if the optional argument var_set is not specified. If it is of interest to evaluate the factor sulphates as a whole, the function surr_rsq_rank() allows us to group sulphates, ${\mathtt{sulphates}^{2}}$, and ${\mathtt{sulphates}^{3}}$ by using the optional argument var_set. For example, in the code below we create a list of two groups: one group contains all terms of sulphates and the second group only contains higher order terms of sulphates.
The output table above shows that the factor sulphates in fact contributes 13.82% to the total surrogate ${R^{2}}$ if its linear, squared, and cubic terms are considered altogether. This percentage contribution is much higher than that when only the linear term of sulphates was evaluated (3.13%). By this result, sulphates is lifted to the second place in terms of its relative contribution to the total surrogate ${R^{2}}$. The output table also shows that if we only consider the higher order terms of sulphates, the percentage contribution is 6.19%, which is higher than any other individual variables except volatile.acidity (7.12%). This is another piece of evidence that can support the inclusion of the squared and cubic terms of sulphates in the full model.
5.2.5 Comparability of the Surrogate ${R^{2}}$ Across Different Samples and Models
One of the motives of [26] is to find an ${R^{2}}$ measure so that we can compare the goodness of fit across different models (e.g., linear, binary, or ordinal regression models) and/or samples that address the same or similar scientific/business question. We use the wine data in [10] to demonstrate that the surrogate ${R^{2}}$ enables this comparability, which may lead to new insights into decision-making. [10]’s data include 1599 red wine samples and 4898 white wine samples. Although the same rating scale (i.e., from 0 to 10) was offered to wine experts, in the red wine sample only 6 rating categories (3 to 8) were observed whereas, in the white wine sample, 7 rating categories (3 to 9) were observed. As a result, the ordered probit models fitted to red and white wine samples have a different number of intercept parameters. In addition, after conducting the same analysis but for the white wine sample (using a similar code as presented before), we find out that the set of selected variables is not the same. The 7 selected variables are alcohol, volatile.acidity, residual.sugar, free.sulfur.dioxide, sulphates, fixed.acidity, and pH. As a result, the ordered probit models fitted to red and white wine samples have a different number of slope parameters as well. Given the differences between the samples and models, the surrogate ${R^{2}}$, nevertheless, enables us to compare goodness-of-fit measures across the board. Table 2 summarizes the result obtained using our developed package $\mathbf{SurrogateRsq}$.
Table 2
Percentage contributions and ranks of the physicochemical variables in the analysis of the red wine and white wine samples.
| Red wine data | White wine data | ||||
| Surrogate ${R^{2}}=0.439$ | Surrogate ${R^{2}}=0.307$ | ||||
| Variable | Contribution | Ranking | Contribution | Ranking | |
| alcohol | 25.80% | 1 | 77.16% | 1 | |
| sulphates (& higher-order terms) | 13.82% | 2 | 0.51% | 5 | |
| volatile.acidity | 7.12% | 3 | 20.39% | 2 | |
| total.sulfur.dioxide | 3.52% | 4 | |||
| pH | 2.78% | 5 | 0.06% | 7 | |
| chlorides | 1.21% | 6 | |||
| free.sulfur.dioxide | 0.96% | 7 | 1.42% | 4 | |
| residual.sugar | 5.34% | 3 | |||
| fixed.acidity | 0.32% | 6 | |||
| sulphates${^{2}}$ & sulphates${^{3}}$ | 6.19% | ||||
By comparing the result in the two panels (red versus white wine) of Table 2, we can make the following conclusions: (i) the same set of measured physicochemical features in the experiment of [10] has greater explanatory power for red wine (43.9% versus 30.7%); (ii) the ranking of explanatory variables is different for the two types of wine with only one exception which is alcohol (top for both); and (iii) the percentage contributions of each variable differ significantly in magnitude for red versus white wine (e.g., alcohol, 25.80% versus 77.16%; sulphates, 13.82% versus 0.51%; volatile.acidity, 7.12% versus 20.39%). These insights drawn from our goodness-of-fit analysis may be useful to help us understand how physicochemical features influence wine ratings and how the influence may be different depending on the type of wine. The percentage contributions and ranking of physicochemical features may be used to guide or even devise the wine-making process.
6 Discussion
In this paper, we have developed the R package $\mathbf{SurrogateRsq}$ for categorical data goodness-of-fit analysis using the surrogate ${R^{2}}$. The package applies to probit/logistic regression models, and it is compatible with commonly used R packages for binary and ordinal data analysis. With $\mathbf{SurrogateRsq}$, we are able to obtain the point estimate and the interval estimates of the surrogate ${R^{2}}$. An importance ranking table for all explanatory variables can be produced as well. These new features can be used in conjunction with other R packages developed for variable selection and model diagnostics. This “whole-analysis” is summarized in a workflow diagram, which can be followed in practice for categorical data analysis. To examine the utility of this package in real data analysis, we have used a wine rating dataset as an example and provided the sample codes. In addition, we have used the package $\mathbf{SurrogateRsq}$ to demonstrate that the surrogate ${R^{2}}$ allows us to compare different models trained from the red wine sample and white wine sample. The comparison has led to new findings and insights that deepen our understanding of how physicochemical features influence wine quality. The result suggests that our package can be used in a similar way to analyze multiple studies (and/or models) that address the same or similar scientific or business question.
We use the red wine data to examine the computational time of the functions in our package. Table 3 presents the comparison, where the column (n = 1597) corresponds to the real data, and the other columns (n = 3000, 6000, 12000) are based on pseudo-real data sets generated by randomly sampling more rows from the real data. The numbers are the average running time in seconds over 10 times of repetition on an Apple Macbook Pro Max with the M1 Max CPU. The upper panel of Table 3 shows if only a point estimate of the surrogate ${R^{2}}$ is needed, our surr_rsq() function takes almost no time to provide the result (e.g., merely 0.213 seconds when $n=12000$). However, the bottom panel of Table 3 shows if confidence intervals are wanted, our surr_rsq_ci() function takes longer time with one core of CPU (e.g., 1421 seconds = 23.6 minutes when $n=12000$). Given that a CPU with 6 10 cores is quite common nowadays, we recommend using parallel computing aforementioned. It reduces the computing time to 211.93 seconds = 3.5 minutes when $n=12000$.
Table 3
Computational time estimates of the functions in SurrogateRsq package. The presented numbers in the table are the average time in seconds over 10 times of repetition for these scenarios using an Apple M1 Max Chip with 10 cores and a clock rate of 2.06 ∼ 3.22 GHz. Note: The asym and parallel options are the arguments controlling the asymptotic version of surrogate ${R^{2}}$ and the parallel computing introduced in Section 3.
| Function: surr_rsq() | n=1,597 | n=3,000 | n=6,000 | n=12,000 |
| avg.num = 30 | 0.048 | 0.067 | 0.123 | 0.213 |
| asym = TRUE | 0.001 | 0.002 | 0.004 | 0.007 |
| Function: surr_rsq_ci(B = 2000) | ||||
| avg.num = 30, parallel = FALSE | 241.95 | 403.73 | 777.29 | 1421.41 |
| asym = TRUE, parallel = FALSE | 149.40 | 258.97 | 528.27 | 977.23 |
| avg.num = 30, parallel = TRUE | 35.68 | 60.17 | 116.40 | 211.93 |
| asym = TRUE, parallel = TRUE | 21.54 | 35.85 | 118.11 | 144.95 |
If software developers want to build or modify this package for their specific scientific inquiries, they can modify one or all of the three components of our package. First, what we really need as an input for the functions in our $\mathbf{SurrogateRsq}$ package is the fitted model from another model training package (e.g., glm(), polr()). Software developers can replace the object with the model of their interest. For example, this surrogate ${R^{2}}$ approach may still work for the discrete choice models studied by [8]. Second, depending on the form of the model, software developers can choose or modify what distribution to use for simulating the surrogate response. Third, once the surrogate responses are available, one can follow the inference procedures discussed in our paper and tailor them to meet specific needs.